Some time ago, I put together a proof of concept that that outlined a method that could be utilized to convert the JSON-LD data made available as part of the Homosaurus vocabulary to MARC for use in MARC-centric systems. The original post can be found here: https://blog.reeset.net/archives/2953
In the time since that post, the vocabulary has created a new version of the vocabulary, and I’ve been getting periodic questions from folks, looking for a more simplified method of generating and managing this vocabulary long-term for use in their library catalogs. Since I was already doing some work in MarcEdit to update a couple of integrations, I decided that it would likely be easiest to create a plugin that could simplify the generation process.
If you’d like to see how the plugin works — you can find it here: https://youtu.be/dzZSjj6e9nc?si=9SagrvLyODpxhjFM
This video demonstrates how the plugin will work. Essentially, I tried to simply the process so that it could be run by just clicking on two buttons — one select a save path, and one to start the process.
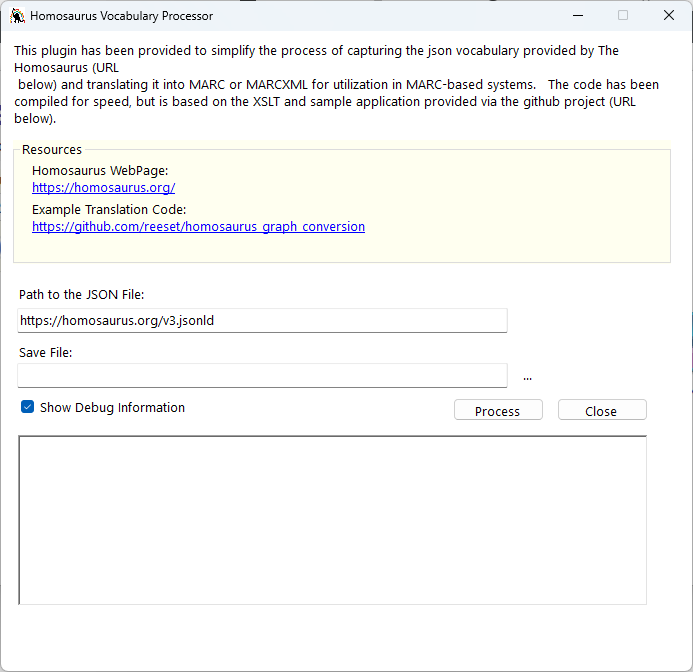
My hope was that by making the interface easy — anyone will be able to utilize the tool with minimal effort. However, knowing catalogers — there will also be a desire to have some level of control and customization over how the records are generated. In the proof-of-concept code, this was done by providing users with the XSLT utilized to generate the MARCXML records. The challenge here is that it assumed that folks would be familiar with the XSLT processes I was using — including the inline inclusion of two C# functions that were create as a kindof – inline extension — to the XSLT processor. I’ll continue to make this code available, but for the plugin, I wanted to simply the process for users…and hopefully I have.
In the plugin, users will be able to control what information populates into the generated subject authority records by accessing a set of template files. These are found in the application configuration direction/plugins75.
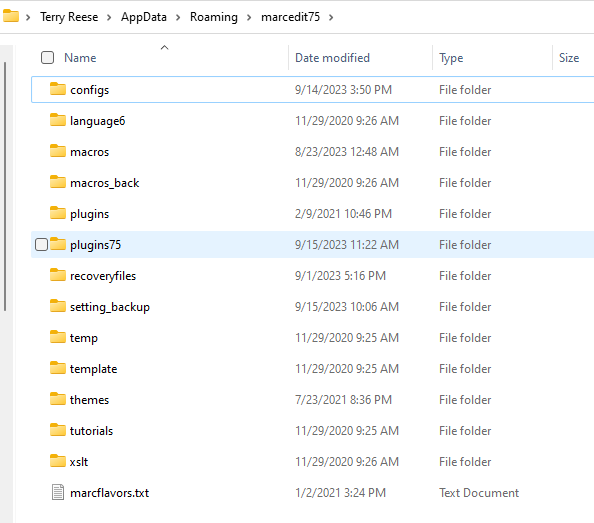
Within the plugins folder, there is a set of templates for the vocabulary:
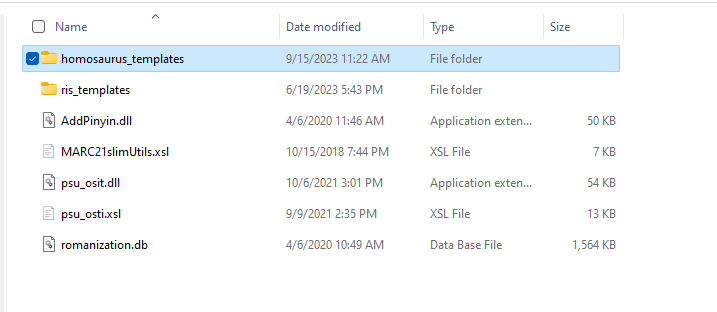
Inside this template folder are a set of XML fragments that define a record template and the templates for each field to be generated.
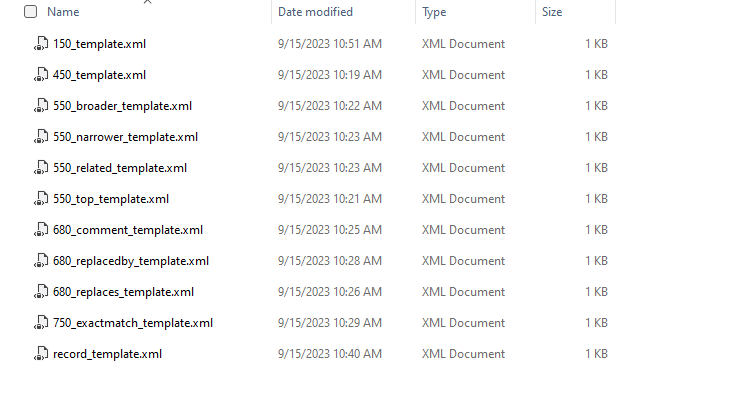
In reviewing the vocabulary — a couple of assumptions are made.
- All subjects are generated as topic headings. This is because the vocabulary doesn’t denote the type of heading being created. So, all authority records will utilize a 150.
- The vocabulary denotes a number of relationships specifically:
– has top container
– broader term
– narrower term
– related term
– comments
– replaced by references
– replaces references
– exact match uris with id.loc.gov - To account for each of these relationships, a template with a field fragment has been created. For example — for narrower terms the following fragment is used:
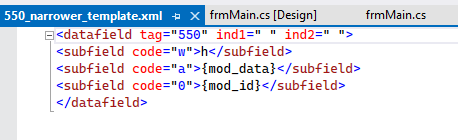
Each fragment includes mnemonics that denote data being pulled from the vocabulary and resolved by the application. In this case, that information goes into the $a (the preferred label) and $0 (the uri). Additionally, the template for narrower terms includes a $w set to h — which denotes the relationship between the term in the 550 field and the 150 field. In this case, the relationship being represented, is that the term in the 550 represents a narrower concept.
Users have the ability to modify the record template
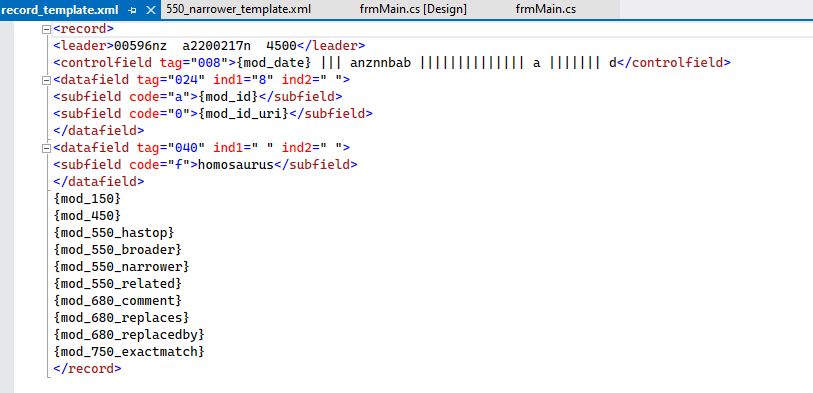
So that folks can refine the leader, 008 or include local information in the 040 – as well as include local data in the record. Users can also decide to not include some data in the outputted MARC/MARCXML records by simply removing the mnemonic from this file. For example, if a user didn’t want to include the 750 reference noting an exact match to a specific term within LCSH – a user would simply need to remove {mod_750_extractmatch} from the record template.
By utilizing this templating method — users that make no changes can expect to see the generation of records that look something like this:
=LDR 04887nz a2200757n 4500 =008 211208\|||\anznnbab\||||||||||||||\a\|||||||\d =024 8\$ahomoit0000669$0https://homosaurus.org/v3/homoit0000669 =040 \\$fhomosaurus =150 \\$aIntersex variations =450 \\$aIntersex conditions =550 \\$wg$aIntersex variations$0https://homosaurus.org/v3/homoit0000669 =550 \\$wg$aIntersex$0https://homosaurus.org/v3/homoit0000352 =550 \\$wg$aMedicalization$0https://homosaurus.org/v3/homoit0001006 =550 \\$wh$aAndrogen insensitivity syndrome$0https://homosaurus.org/v3/homoit0000044 =550 \\$wh$aAphallia$0https://homosaurus.org/v3/homoit0000064 =550 \\$wh$aClitoromegaly$0https://homosaurus.org/v3/homoit0000283 =550 \\$wh$aCongenital adrenal hyperplasia$0https://homosaurus.org/v3/homoit0000301 =550 \\$wh$aGonadal dysgenesis$0https://homosaurus.org/v3/homoit0000599 =550 \\$wh$aHypospadias$0https://homosaurus.org/v3/homoit0000657 =550 \\$wh$aKlinefelter syndrome$0https://homosaurus.org/v3/homoit0000692 =550 \\$wh$aMicropenis$0https://homosaurus.org/v3/homoit0001014 =550 \\$wh$aMosaicism involving "sex" chromosomes$0https://homosaurus.org/v3/homoit0001020 =550 \\$wh$aOvo-testes$0https://homosaurus.org/v3/homoit0001066 =550 \\$wh$aPartial androgen insensitivity syndrome$0https://homosaurus.org/v3/homoit0001079 =550 \\$wh$aProgestin induced virilization$0https://homosaurus.org/v3/homoit0001134 =550 \\$wh$aSwyer syndrome$0https://homosaurus.org/v3/homoit0001354 =550 \\$wh$aTurner syndrome$0https://homosaurus.org/v3/homoit0001478 =550 \\$wh$a5-alpha reductase deficiency$0https://homosaurus.org/v3/homoit0000001 =550 \\$wh$aEpispadias$0https://homosaurus.org/v3/homoit0002836 =550 \\$wh$a17-beta hydroxysteroid dehydrogenase 3 deficiency$0https://homosaurus.org/v3/homoit0002818 =550 \\$wh$aAmbiguous genitalia$0https://homosaurus.org/v3/homoit0002819 =550 \\$wh$aAnorchia$0https://homosaurus.org/v3/homoit0002821 =550 \\$wh$aAromatase deficiency$0https://homosaurus.org/v3/homoit0002822 =550 \\$wh$aAromatase excess syndrome$0https://homosaurus.org/v3/homoit0002823 =550 \\$wh$aBladder exstrophy$0https://homosaurus.org/v3/homoit0002824 =550 \\$wh$aChimerism$0https://homosaurus.org/v3/homoit0002826 =550 \\$wh$aCloacal exstrophy$0https://homosaurus.org/v3/homoit0002828 =550 \\$wh$aComplete androgen insensitivity syndrome$0https://homosaurus.org/v3/homoit0002829 =550 \\$wh$aCongenital chordee$0https://homosaurus.org/v3/homoit0002831 =550 \\$wh$aCryptorchidism$0https://homosaurus.org/v3/homoit0002832 =550 \\$wh$aDe la Chapelle syndrome$0https://homosaurus.org/v3/homoit0002833 =550 \\$wh$aEstrogen insensitivity syndrome$0https://homosaurus.org/v3/homoit0002837 =550 \\$wh$aFollicle-stimulating hormone insensitivity$0https://homosaurus.org/v3/homoit0002838 =550 \\$wh$aFraser syndrome$0https://homosaurus.org/v3/homoit0002839 =550 \\$wh$aHyperandrogenism$0https://homosaurus.org/v3/homoit0002842 =550 \\$wh$aHypogonadism$0https://homosaurus.org/v3/homoit0002843 =550 \\$wh$aIsolated 17,20-lyase deficiency$0https://homosaurus.org/v3/homoit0002848 =550 \\$wh$aJacobs syndrome$0https://homosaurus.org/v3/homoit0002861 =550 \\$wh$aKallmann syndrome$0https://homosaurus.org/v3/homoit0002862 =550 \\$wh$aLeydig cell hypoplasia$0https://homosaurus.org/v3/homoit0002863 =550 \\$wh$aLuteinizing hormone insensitivity$0https://homosaurus.org/v3/homoit0002864 =550 \\$wh$aMayer-Rokitansky-Küster-Hauser syndrome$0https://homosaurus.org/v3/homoit0002865 =550 \\$wh$aMüllerian duct variations$0https://homosaurus.org/v3/homoit0002868 =550 \\$wh$aPenoscrotal transposition$0https://homosaurus.org/v3/homoit0002870 =550 \\$wh$aPersistent Müllerian duct syndrome$0https://homosaurus.org/v3/homoit0002871 =550 \\$wh$aPolycystic ovary syndrome$0https://homosaurus.org/v3/homoit0002872 =550 \\$wh$aPseudohermaphroditism$0https://homosaurus.org/v3/homoit0002873 =550 \\$wh$aSex reversal syndrome$0https://homosaurus.org/v3/homoit0002875 =550 \\$wh$aTrisomy X$0https://homosaurus.org/v3/homoit0002878 =550 \\$wh$aVaginal atresia$0https://homosaurus.org/v3/homoit0002883 =550 \\$wh$aXXYY syndrome$0https://homosaurus.org/v3/homoit0002885 =550 \\$aSex assigned at birth$0https://homosaurus.org/v3/homoit0001268 =550 \\$aAssigned X at birth$0https://homosaurus.org/v3/homoit0001646 =550 \\$aIntersex people$0https://homosaurus.org/v3/homoit0000618 =680 \\$iThis authority record replaces Intersex conditions$0http://homosaurus.org/v2/intersexConditions =750 \0$aIntersexuality$0http://id.loc.gov/authorities/subjects/sh85060401
However, for users interesting in making records their own — this new process will hopefully provide folks with a much more simplified process to support capture and generation of subject authority records for this vocabulary.
Question — feel free to let me know.
–tr
Comments
2 responses to “Simplifying MARC generation of the Homosaurus vocabulary”
Hi Terry,
Thank you for putting this plugin! This is something I’ve been looking for and I’m excited to give it a try, however, I’m not seeing the plug-in listed in my plugin manager. I’m running MARCEdit 7.6.11 on a PC. Is there something I need to download?
Thank you,
Celia
The plugin will show up in 7.6.12+. The work on the plugin is finished, I’m completing updates to the OCLC integration (shifting to the metadata v2 apis). This work is done, but in testing.
–tr